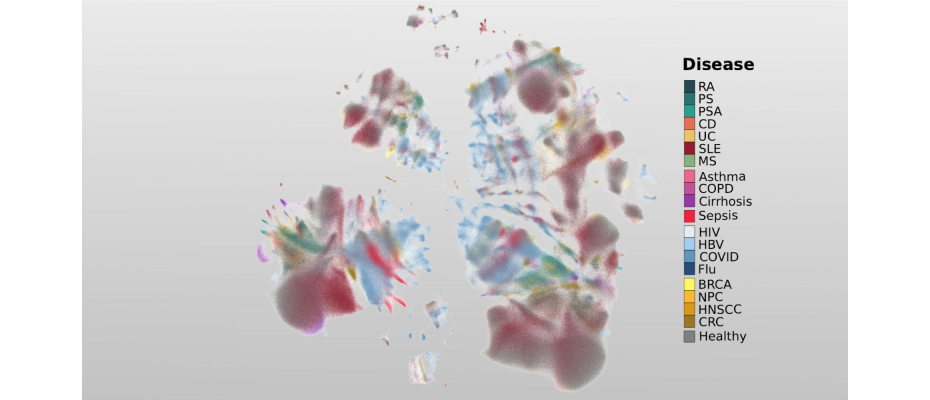
- Researchers from the Centro Nacional de Análisis Genómico (CNAG) have launched the first cellular atlas of inflammation, an exhaustive framework that includes the analysis of more than 6.5 million blood cells from over 1,000 healthy individuals and patients across 19 diseases.
- The comprehensive dataset served as the foundation for developing a biomedical artificial intelligence (AI) tool that learns from cellular immune states implicated in disease mechanisms to extract inflammatory biomarkers, with the aim of supporting future patients in understanding and diagnosing their condition.
- This innovative approach, published in Nature Medicine, lays the groundwork for developing precision diagnostic tools, enabling faster identification of patients with autoimmune and inflammatory diseases, infections, and certain tumours, such as breast or colorectal cancer.
Inflammation is a natural part of everyday life. When you scrape your knee or catch a cold, your body mounts an inflammatory response to restore balance and repair tissues, an essential process for maintaining health. Yet, in many diseases, this response becomes disrupted. How inflammation goes wrong, and how these changes differ across conditions such as infections like COVID-19, immune-related diseases including psoriasis or rheumatoid arthritis, and cancers such as breast or colorectal cancer, remains poorly understood. To shed light on this missing piece, researchers at the Centro Nacional de Análisis Genómico (CNAG), in collaboration with the École Polytechnique Fédérale de Lausanne (EPFL, Switzerland), have created the first cellular atlas of inflammation, a comprehensive resource based on more than 6.5 million blood cells from over 1,000 healthy individuals and patients spanning 19 different diseases.
Published in the prestigious journal Nature Medicine, this study introduces an innovative approach: viewing cells themselves as a biomarker that can help uncover some of the mysteries behind pathological inflammatory conditions. “Immune cells from the blood contain fingerprints of diseases, which they pick up when circulating through our system. Reading them at cellular resolution allows us to convert cells into living biomarkers for precision diagnostics”, explains Dr Holger Heyn, senior and corresponding author of the study and leader of the Single Cell Genomics Group at CNAG. “We provide the proof-of-concept to convert immune cells into a universal diagnostic tool, a concept we are now moving to clinical prototyping stage”.
 The research examines inflammatory signals from immune cells in the blood and what they can reveal about the diseases affecting patients. By analysing in detail different immune cell states that drive disease progression, the study uncovers how inflammation changes across conditions. Using the new cellular atlas, researchers identified molecular indicators that act as signposts, guiding the way toward a deeper understanding of the underlying biology and to precisely classify these diseases. To find these signposts, the researchers zoomed into the inflammatory genes that orchestrate the immune response, identifying programs that activate immune cells, guide their movement, defend against harmful agents, and trigger defence. This genetic insight helps distinguish each disease and classify patients based on their unique inflammatory signatures.
The research examines inflammatory signals from immune cells in the blood and what they can reveal about the diseases affecting patients. By analysing in detail different immune cell states that drive disease progression, the study uncovers how inflammation changes across conditions. Using the new cellular atlas, researchers identified molecular indicators that act as signposts, guiding the way toward a deeper understanding of the underlying biology and to precisely classify these diseases. To find these signposts, the researchers zoomed into the inflammatory genes that orchestrate the immune response, identifying programs that activate immune cells, guide their movement, defend against harmful agents, and trigger defence. This genetic insight helps distinguish each disease and classify patients based on their unique inflammatory signatures.
For example, in systemic lupus erythematosus, an aggressive autoimmune disorder, the maps revealed that certain immune cell states are linked to more severe forms of the condition. One of the key signals involved is the interferon response, a natural defence mechanism that usually helps fight viruses. In lupus, this signal is constantly active, which can overstimulate the immune system and worsen inflammation. This discovery helps researchers understand why some cases become chronic and more aggressive.
From maps to meaning: how AI learns from inflammation
In addition to using the latest single-cell genomics techniques from CNAG, the study introduces a second major innovation: the application of AI to classify patients based on single-cell data. The researchers developed a generative AI model capable of learning from the cellular states and gene activity captured in the atlas, with the goal of projecting the biological patterns to future patients. This new approach lays the groundwork for a precision medicine tool that could accelerate diagnosis and guide more personalised treatments, ultimately improving both the quality of life and the clinical management of individuals affected by diseases with inflammatory conditions.
Dr Juan Nieto, immunologist in the Single Cell Genomics Group at CNAG and co-corresponding author of the study, highlights: “This atlas helps us move toward more personalised care. By understanding the specific inflammatory patterns present in each patient, clinicians may be able to choose treatments earlier and more precisely, reducing trial-and-error and improving patient care.”
Following the launch of this open-source resource, which has already been tested with patient samples as a personalised diagnostic tool, the researchers are now working on the next phase of the project. Their efforts are currently focused on establishing the protocols needed to ensure quality and standardisation, with the goal of building a robust foundation that enables its integration into clinical practice, making it accessible to hospitals as well as healthcare institutions in the future.
The study has been made possible thanks to the data from DocTIS, a European Union-funded multicentric project aimed at improving treatment efficacy in immune-mediated inflammatory diseases, and to the collaboration of a network of national and European hospitals.
IMAGE
CNAG Researchers involved in the study (from left to right) Dr Holger Heyn, Dr Sergio Aguilar, Dr Ginevra Caratù, Dr Laura Jiménez, Dr Davide Maspero and Dr Juan Nieto.
REFERENCE ARTICLE
Jiménez-Gracia, Laura, et al. ‘Interpretable Inflammation Landscape of Circulating Immune Cells’. Nature Medicine, Jan. 2026, pp. 1–12. www.nature.com, https://doi.org/10.1038/s41591-025-04126-3.











