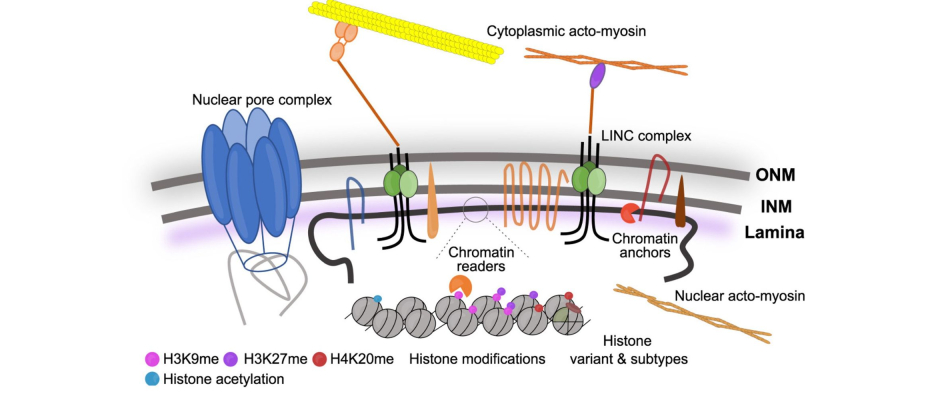
Although it occurs at a scale far smaller than a nanometre, the three-dimensional (3D) organisation of the genome inside each cell is remarkably intricate. One of its key and quite unknown components are the Lamina-Associated Domains (LADs): regions of the genome that interact with the nuclear lamina – the dense fibrous network lining the inside of the nucleus – which not only protects DNA from potential damage but also plays an essential role in gene regulation and cellular function. Understanding how this structure, which is considered the earliest feature of nuclear organization following fertilisation, emerges during the very first stages of development is crucial for advancing our knowledge of genetics and developmental biology.
A study recently published in Cell, titled “The establishment of nuclear organization in mouse embryos is orchestrated by multiple epigenetic pathways”, reveals new insights into this process, and the main epigenetic pathways that contribute to gene regulation. The research was led by the Institute of Epigenetics and Stem Cells in Munich (Germany) in collaboration with CNAG and our Structural Genomics Group, headed by Prof. Marc A. Marti-Renom. CNAG contributed particularly to the computational analysis of the data, helping uncover the mechanisms behind genome organisation in the earliest embryonic stages.
The study explores how epigenetic factors – chemical and structural modifications that do not alter the DNA sequence but regulate how it functions – drive the formation of the nuclear architecture in mouse zygotes and 2-cell-stage embryos, using in vivo approaches. The research identifies several pathways that contribute to the organisation of the genome’s 3D structure. These include changes in histone H3, a protein that regulates gene activity, modifications in heterochromatin, which helps silence certain genes, and the histone content, or the amount of these proteins present, which influences how the genome is packaged and organised within the nucleus.
Among the key findings, the researchers discovered that embryos have an incredible ability to rebuild the LAD architecture by the time they reach the 2-cell stage. Even when this structure is disrupted at the zygote stage, the embryo can quickly and effectively reorganize. This shows the resilience and remarkable organizational capacity of early embryos, almost as if they have a built-in blueprint for rebuilding themselves.
The study also reveals that maternal inheritance of the H3K27me3 marker, which is associated with gene repression, plays a key role in shaping the formation of LADs in the zygote. This epigenetic marker acts as a switch, determining whether a gene should be silenced or active. Additionally, the research demonstrates that LAD boundaries reorganise based on positional information provided by histone marks such as H3K4me3, which is linked to active genes, and H3K9me3, associated with repressed genes. These marks guide the repositioning or reorganisation of LADs, helping define the active and silent regions of the genome.
This research opens new avenues for understanding how the genome’s 3D structure forms from the earliest stages of development and provides crucial insights into the epigenetic processes that regulate gene expression. The findings also hold significant implications for our understanding of early development and genetic regulation.
REFERENCE ARTICLE
Pal, Mrinmoy, et al. «The Establishment of Nuclear Organization in Mouse Embryos Is Orchestrated by Multiple Epigenetic Pathways». Cell, abril de 2025, p. S0092867425003964. DOI.org (Crossref), https://doi.org/10.1016/j.cell.2025.03.044.











